Welcome to RBP2GO 2.0!
A comprehensive pan-species resource on RNA-binding proteins, their interactions and functions
Quick start
Search Proteins
This search option is not case sensitive.
To search for multiple proteins (e.g. copy/paste a protein list), use the "Advanced Search" option.
Press enter or the search icon on the right to start the search. A list of proteins will be computed.
Search Gene Ontology
This search option is not case sensitive.
To search for multiple GO terms, use the "Advanced Search" option.
Press enter or the search icon on the right to start the search. A list of proteins will be computed.
Search Disease Ontology
This search option is not case sensitive.
The exact DO term or DO ID needs to be provided, a keyword search with e.g. "breast" or "Cancer" does not work
To search for multiple DO terms or to find DO terms use the Advanced Search option and its help menu (folder-tree icon on the right)
Press enter or the search icon on the right to start the search. A list of proteins will be computed.
Search InterPro Features
For multiple entries, enter each term on a separate line.
The RNA-binding status is only available for InterPro Families, Domains and Repeats according to Wassmer et al. 2024 .
The sign "-" in the column "RNA-binding" refers to the abscence of information with regard to the RNA-binding status of the feature.
In the column "->", if "Search Proteins" is clicked, it sends the InterPro feature ID to the "Advanced Search".
Press the search icon on the right to start the search. A list of InterPro features will be computed.
How to cite
Cantarella, Neffe et. al. RBP2GO 2.0: Integrating Disease Associations and Sequence Features to explore RNA-Binding Protein Functions. Nucleic Acids Research, gkaf1258, 2025General Statistics
Species-specific statistics
RBP2GO Publications
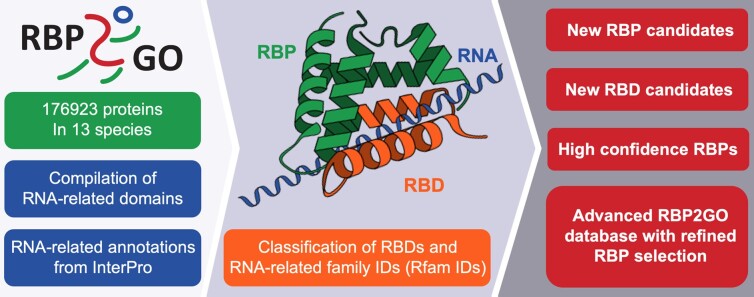
Search RBP2GO by proteins, Gene Ontology or Disease Ontology
Use this section to perform individual searches by protein name, Gene Ontology, or Disease Ontology.
Only one type of search is allowed at a time.
The result of the search is a table of proteins, which are displayed alphabetically by species. Within each species, the proteins are sorted in descending order according to the RBP2GO Composite Score of the proteins (about scores in RBP2GO).
The search options can be combined within Advanced Search, which provides additional parameters in a flexible and user friendly manner.
Search Proteins
This search option is not case sensitive.
To search for multiple proteins (e.g. copy/paste a protein list), use the "Advanced Search" option.
Press enter or the search icon on the right to start the search. A list of proteins will be computed.
Search Gene Ontology
This search option is not case sensitive.
To search for multiple GO terms, use the "Advanced Search" option.
Press enter or the search icon on the right to start the search. A list of proteins will be computed.
Search Disease Ontology
This search option is not case sensitive.
The exact DO term or DO ID needs to be provided, a keyword search with e.g. "breast" or "Cancer" does not work
To search for multiple DO terms or to find DO terms use the Advanced Search option and its help menu (folder-tree icon on the right)
Press enter or the search icon on the right to start the search. A list of proteins will be computed.
Protein Info
Its values ranges from 0 to 100.
The RBP2GO Composite Score builds on the RBP2GO score and combines it with a domain score based on the presence of a known RNA-binding InterPro domain, repeat or family.
The violin plots depict the distributions of the RBP2GO Score and BRP2GO Composite Score in the given species. The median indicated as a white dot. The score of the selected protein is marked by the vertical line.
A comprehensive description of the RBP2GO Score and RBP2GO Composite Score is available here
Its values ranges from 0 to 100.
The RBP2GO Composite Score builds on the RBP2GO score and combines it with a domain score based on the presence of a known RNA-binding InterPro domain, repeat or family.
The violin plots depict the distributions of the RBP2GO Score and BRP2GO Composite Score in the given species. The median indicated as a white dot. The score of the selected protein is marked by the vertical line.
A comprehensive description of the RBP2GO Score and RBP2GO Composite Score is available here
Cancer Gene Census Details
Disease Involvement
RBP Studies
Studies:
Sequence Features
RBDs:
Download TableOther InterPro annotations:
Download TableGene Ontology
Homology
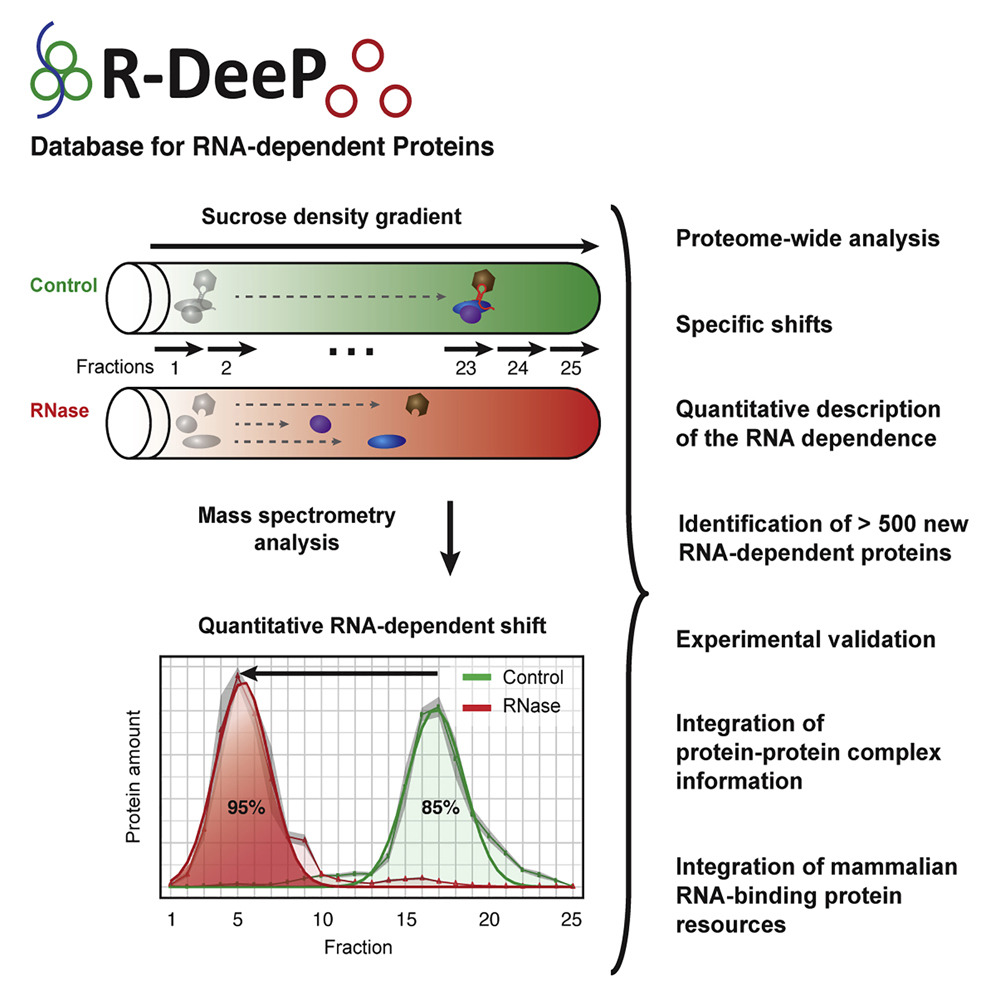
R-DeeP
R-DeeP datasets were collected in different cell types and different phases of the cell cycle.Disease Ontology
Confident Disease Ontology Terms
Download TableAll Disease Ontology Terms
Download TableSearch InterPro Features
Use this section to perform
individual or multiple searches for InterPro features
such as protein domains, repeats or families.
We refer here only to terms and IDs from the InterPro database.
RNA-binding domains, repeats and families were classified
according to
1- three experimental studies:
Castello et al. 2012
;
Gerstberger et al. 2014
;
Castello et al. 2016
2- a keyword search from the InterPro database
3- the frequency of the InterPro features (domains, repeats and families) in the proteins experimentally identified as RBPs.
For a
detailed description of the procedure,
see
Wassmer et al. 2024
.
Search InterPro Features
For multiple entries, enter each term on a separate line.
The RNA-binding status is only available for InterPro Families, Domains and Repeats according to Wassmer et al. 2024 .
The sign "-" in the column "RNA-binding" refers to the abscence of information with regard to the RNA-binding status of the feature.
In the column "->", if "Search Proteins" is clicked, it sends the InterPro feature ID to the "Advanced Search".
Press the search icon on the right to start the search. A list of InterPro features will be computed.
Data & Downloads
General Data
Protein Data
Additional Resources
Species Specific Databases
Proteins and Protein-Protein Interactions Databases
Intrinsically Disordered Proteins and Low Complexity Domains
Coiled-coil Prediction
Gene Ontology
Diseases
Isoelectric point
Databases Collections
Literature Search
Icons and Webpage
About RBP2GO
By offering flexible research options, RBP2GO provides users with opportunities to browse RBPs (via the protein search option) and explore their implications in cell function (via the gene ontology (GO) search option) and disease (via the disease ontology (DO) search option).
Additionally, protein sequence features can be explored with regard to their association with RNA.
The datasets are available for download from the References & Data section.
For more information, please refer to the FAQ section below.
RBP2GO is managed by the group RNA-Protein Complexes and Cell Proliferation at the German Cancer Research Center (DKFZ) in Heidelberg, Germany.
How to cite
Cantarella, Neffe et. al. RBP2GO 2.0: Integrating Disease Associations and Sequence Features to explore RNA-Binding Protein Functions. Nucleic Acids Research, gkaf1258, 2025FAQ
RBP2GO can be used to efficiently explore new avenues in RNA biology and initiate research on unexpected functions of RNA-protein interactions in the cell.
For each protein, the database enables users to explore:
- the large protein resource to extract RBPs and/or Non-RBPs
- the biological functions of the protein
- the associations with human diseases (specific for Homo sapiens)
- the protein sequence features
- the versatile "Advanced Search", which allows combining multiple search parameters with intuitive filtering options
- the interaction partners
- the protein homologs
- the list of available experimental datasets for RBP screening studies (including a link to the publication)
- the available R-DeeP datasets with the respective R-DeeP graphs (specific for Homo sapiens)
- the available datasets for the interacting RNA species
- the RBP2GO Score
- the RBP2GO Composite Score
- a graphical representation (violin plot) positions the score of the protein as compared to the other scores of all proteins or RBPs of the species
However, according to our study Wassmer et al. 2024 the RBP aspirants contain RNA-binding domains (RBDs).
RBP aspirants can be identified in RBP2GO by using the "Advanced Search" module and combining the following options: "Select RBP - Only Non-RBPs" and "Only RBD-containing proteins". This search can also be species specific by using the option "Select Species".
In total, 10420 RBP aspirants are listed in RBP2GO 2.0, for which additional information is provided in the specific protein page.
- Three options are offered in green boxes, for which the searches return a protein list as a result. These options are not case sensitive:
- "Search Proteins": Enter a protein name, UniProt ID or a keyword (e.g., "HNRNPU", "Q00839", or "heterogeneous"). To search for multiple proteins (e.g. copy/paste a protein list), use the "Advanced Search" option. Press enter or the search icon on the right to start the search. A list of proteins will be computed and displayed under the "Search RBP2GO" top panel.
- "Search Gene Ontology": Enter a Gene Ontology (GO) term or a keyword (e.g., "GO:0003723", or "RNA binding"). The GO IDs must start with "GO:". To search for multiple GO terms, use the "Advanced Search" option. Press enter or the search icon on the right to start the search. A list of proteins will be computed under the "Search RBP2GO" top panel.
- "Search Disease Ontology": Enter a Disease Ontology (DO) term (e.g., "DOID:1324" or "lung cancer") or ID. The DO ID must start with "DOID:". The exact DO term or DO ID needs to be provided, a keyword search with e.g. "breast" or "Cancer" does not work. To search for multiple DO terms or to find DO terms use the "Advanced Search" option and its help menu (folder-tree icon on the right). Press enter or the search icon on the right to start the search. A list of proteins will be computed under the "Search RBP2GO" top panel.
- "Search InterPro Features": Enter an InterPro ID or a keyword to search for protein domains, repeats or families (e.g., "IPR025223", "S1-like RNA binding domain"). The InterPro IDs must start with "IPR". For multiple entries, enter each term on a separate line. The RNA-binding status is only available for InterPro Families, Domains and Repeats according to Wassmer et al. 2024 . Press the search icon on the right to start the search. A list of InterPro features will be computed.
The orange search box is dedicated to InterPro features:
The Advanced Search allows users to search for multiple protein names, GO terms, or DO terms, and to filter results by various criteria.
For example, users can:
- restrict the search to a specific species
- include only proteins detected as RNA-binding or RNA-dependent
- limit results by composite score or RBP2GO score
- display only proteins related to Cancer Gene Census (CGC) genes
- show only proteins containing an RBD or a family associated to RNA binding
- for the "Disease Ontology" options, a tree view is provided to help users navigate and find relevant diseases
The content of the downloadable table will be adjusted accordingly.
You can type in any keywords for string-type columns or select ranges for numerical columns. Logical columns can be filtered using "yes" or "no" (see next point).
1- three experimental studies: Castello et al. 2012 ; Gerstberger et al. 2014 ; Castello et al. 2016 .
2- a keyword search from the InterPro database.
3- the frequency of the InterPro features (domains, repeats and families) in the proteins experimentally identified as RBPs.
For a detailed description of the procedure, see Wassmer et al. 2024 .
Importantly, the 15 new RBDs added since the RBP2GO-2-Beta version have been incorporated into the calculation of the RBP2GO Composite Score.
Importantly, the datasets from Bae et al. 2020 and Bae et al. 2021 report only the position of the cross-linked amino acid. The corresponding theoretical cross-linked tryptic peptide was derived through our own calculations using the cleaver R package simulating trypsin digestion sites closest to the crosslinked amino acid. Trypsin was used in the aforementioned studies to generate peptides and identify the crosslinked sites.
- Potential RBPs can be detected within the RBP aspirants, i.e. proteins with RNA-related InterPro features, but which have not yet been detected in proteome-wide RBP screens.
- Non-validated RBPs can be detected within the list of RBPs detected in the proteome-wide screens. In the "Advanced Search" search, filter for the RBPs/R-DeeP proteins. The tool returns a list of candidate proteins sorted by species and then by the RBP2GO Composite Score. Look for the non-validated RBPs.
- Users can detect interesting non-validated or novel RBPs by using our powerful reverse "Gene Ontology" search tools. The gene ontology resource can be browsed according to keywords and returns a list of proteins, which can be RBPs or not. This tool is especially interesting when exploring biological functions and cellular pathways which are not expected to be related to RNA.
Species-specific statistics
RBP2GO Publications



RBP2GO versions
RBP2GO Version 2.0:
release date September 2025
Main change: addition of disease ontology information, RNA-binding peptides and linked to sequencing of the interacting RNAs when available.
RBP2GO 2.0 Beta Version:
release date August 2023
Main change: addition of the RNA-binding domain information and domain search.
Creation of the RBP2GO composite score, based on the RBP2GO score with the inclusion of the RNA-binding domain and
RNA-related InterPro family knowledge.
RBP2GO Version 1.0: release date July 2020
Contact
The authors of the RBP2GO database can be contacted at the following email address: database.RBP2GO@dkfz.de